BIODICA
An integrated computational environment for application of Independent Component Analysis (ICA) to the bulk and single cell molecular profiles, interpretation of the results in terms of biological functions and associations with metadata.
What is BIODICA ?
BIODICA is a computational environment designed for the application of Independent Component Analysis (ICA) to large omics data sets and interpreting the results in a user-friendly manner. It provides a rich set of tools to visualize the results, interpret them using prior knowledge and sample annotations, and perform meta-analyses with several data sets. It comes with the python package stabilized-ica which provides several ICA algorithms, a stabilization procedure to extract reliable and reproducible components, as well as python implementations of several analysis tools proposed by BIODICA. BIODICA is also equipped with a user-friendly graphical user interface allowing non-experienced users to perform the ICA-based analysis on their own transcriptomic data sets.
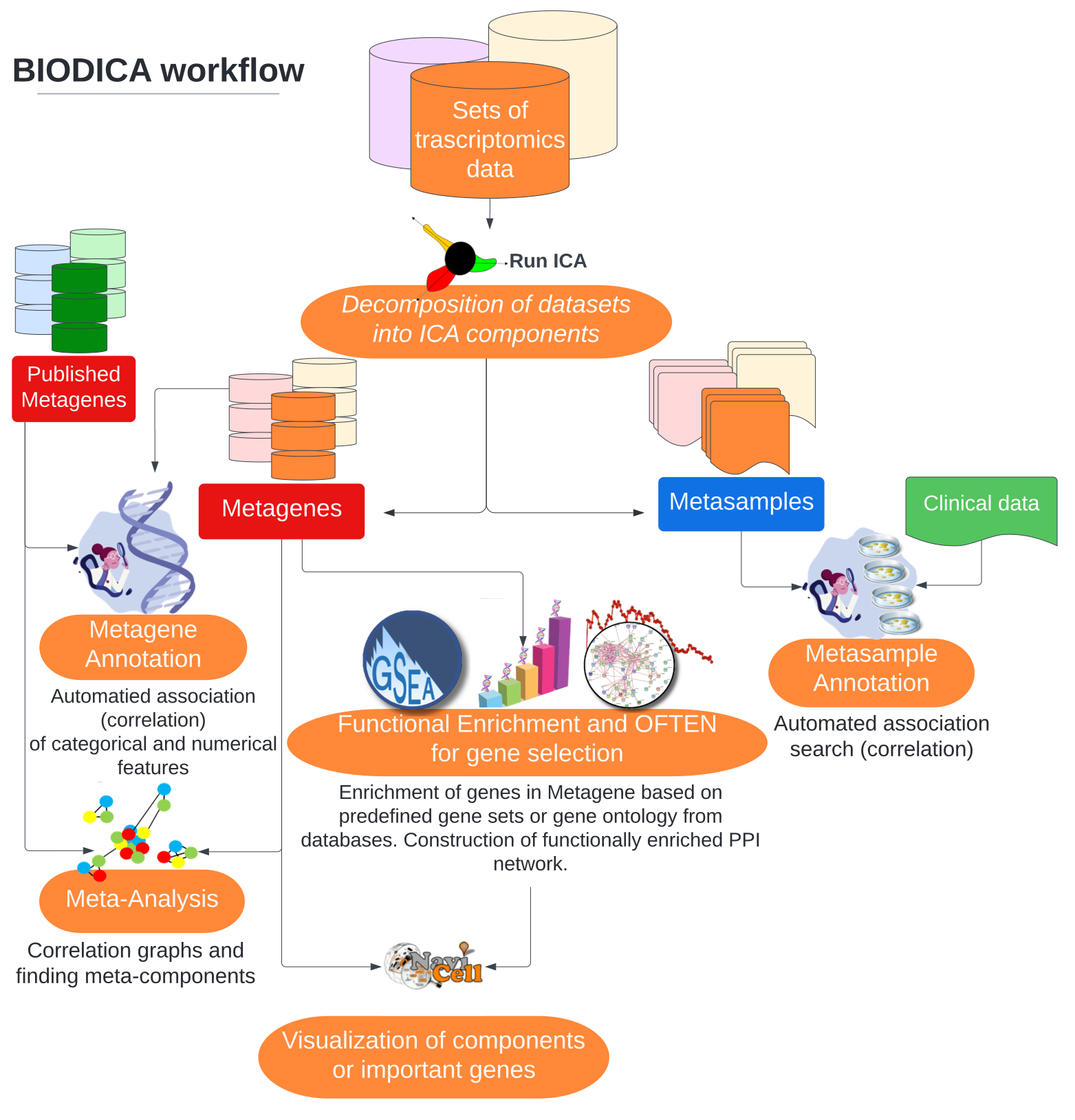

stabilized-ica is the computational core of BIODICA. It can also work as a standalone python tool, facilitating the integration of the ICA-based analysis into complex python pipelines which deal with transcriptomic data. It is available on PyPI and GitHub. Learn more...
